Data Downloading¶
You need to prepare raw data and make a directory for data reduction, before you start data reduction with HSC pipline. There are following 4 steps:
Data downloading¶
You can download your HSC raw data through STARS (Subaru Telescope Archive System) . Please login STARS with your account and password. Once you login, please select Proposal_ID Instrument or Proposal_ID in Search by tab (shown at top left).
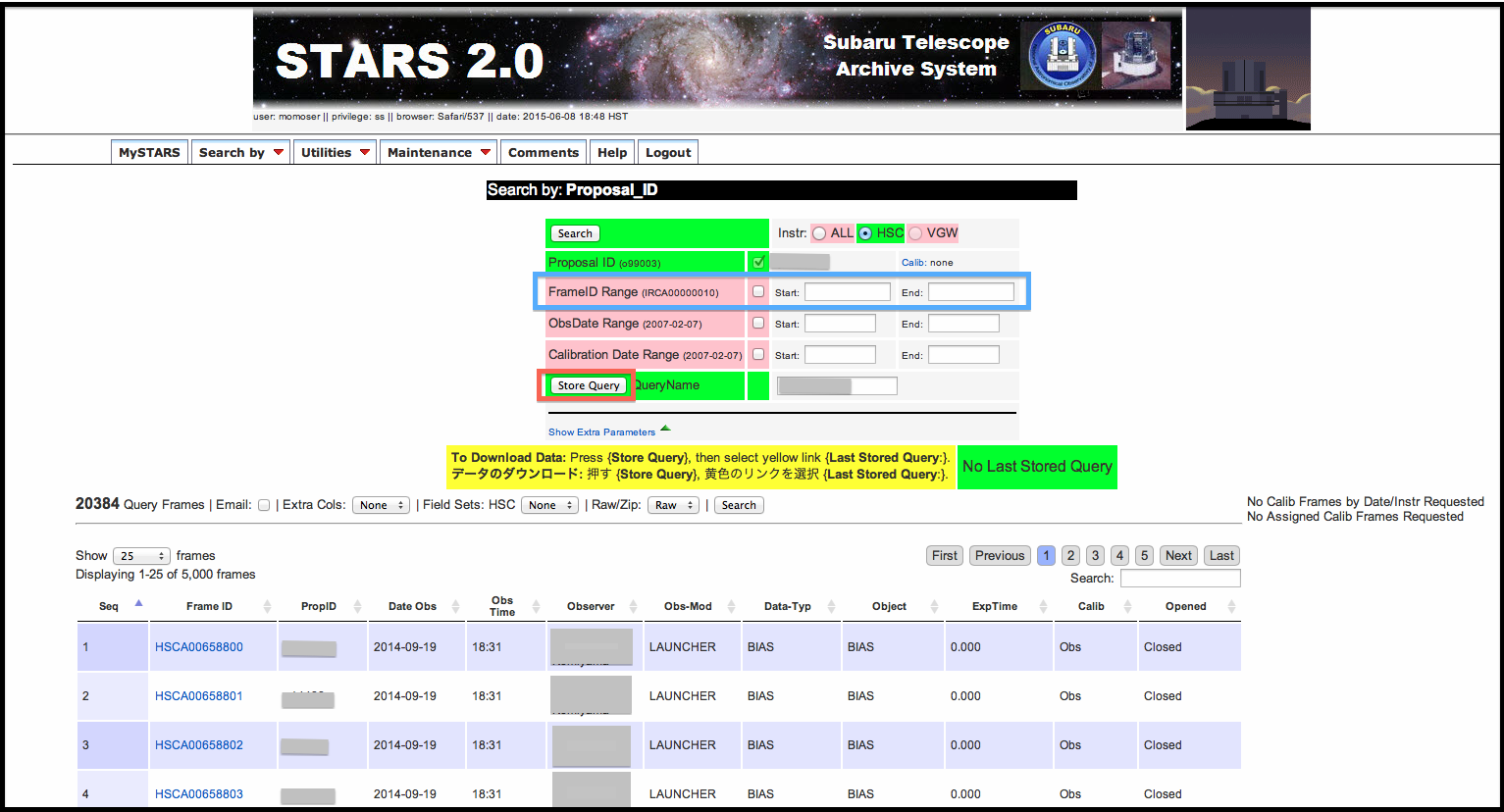
Figure 2 shows search results by Proposal_ID. You can find all HSC raw data of your proposal ID by pressing [search] button. If you want to download all data taken with your proposal ID, you just press [Store query] (a red box) and submit a request. If you only get some specific data, please input raw data ID of the specific data in a blue box and submit a request. It takes some time depending on data amount to be displayed the results of data request.
We show the results of data request in Figure 3. There is a box labeled Wget Input Files at the bottom left. Once you find the box, please download and expand S2Query.tar file into your computer. The python script in S2Query.tar is a script for downloading HSC raw data to your computer. We show an example to download from STARS.
# making a directory to store data
mkdir ~/data
cd data
mv ~/Downloads/S2Query.tar .
# expanding S2Query.tar file
tar xvf S2Query.tar
# Once you expand the tar file,
# zadmin/ zip/ zvalidate/ directories are generated in ~/data directory
# Runnig the python script to download your data
./zadmin/unpack.py
# You must run the script at ~/data direcroty,
# otherwise you are going to fail data downloading.
After you download your data, please check your data directory (~/data) whether there are fits files named HSCA123456**.fits. Note that we introduce the naming method for HSC data in Naming HSC data.
Setup HSC pipeline¶
Please set up HSC pipeline after data downloading. You must set up HSC pipeline for every time you login your computer.
cd ~/
# a setup command of HSC pipeline
setup-hscpipe
Make a directory for data reduction¶
You make a repository which is a work-directory for your data reduction, and whose structure is shown in Usage of HSC pipeline. It is very important to create _mapper file in a repository because the _mapper file indicates the location of your data.
# making a repository
mkdir ~/hsc
# creating a _mapper file
echo 'lsst.obs.hsc.HscMapper' > ~/hsc/_mapper
Set a registry for raw data¶
You need to move your raw data into a repository, and register the data to HSC pipeline by hscIngestImages.py command.
# moving raw data into a repository
cd ~/data
# registering raw data to HSC pipeline
hscIngestImages.py ~/hsc ./*.fits --mode=link --create
# USAGE: hscIngestImages.py <data reduction directory> <name of fits file> --mode=*** --create
#
# Parameters:
# --mode : Raw data placement method when you make a new registry. Options are move, copy, link, and skip.
# --create: A parameter when you make a new registry.
Warning
Please specify a directory with absolute path when you use HSC pipeline commands.
Once you set a registry with hscIngestImages.py, new directories of <object name>, DOMEFLAT, DARK and so on are generated under ~/hsc. Additionally, you also find your HSC raw data in <object name> directory. Information of raw data is registered in a file named registry.sqlite3 under ~/hsc. You can retrieve the information in registry.sqlite3 by SQL commands.
We introduce data reduction of G-band and I-band data of DITH-16H as an example. First, we retrieve information in registry.sqlite3.
# opening registry.sqlite3 by SQL
sqlite3 ~/hsc/registry.sqlite3
# reading a header information
sqlite> .header on
# selecting visit ID, filter name, field name, number of visits
sqlite> SELECT visit, filter, field, count(visit)
...> FROM raw # selecting above information from a registry
...> GROUP BY visit, filter, field; # showing information in the order of visit > filter > field
# (Search results are displaied as Figure 4)
# quit retrieving a registory
sqlite> .q
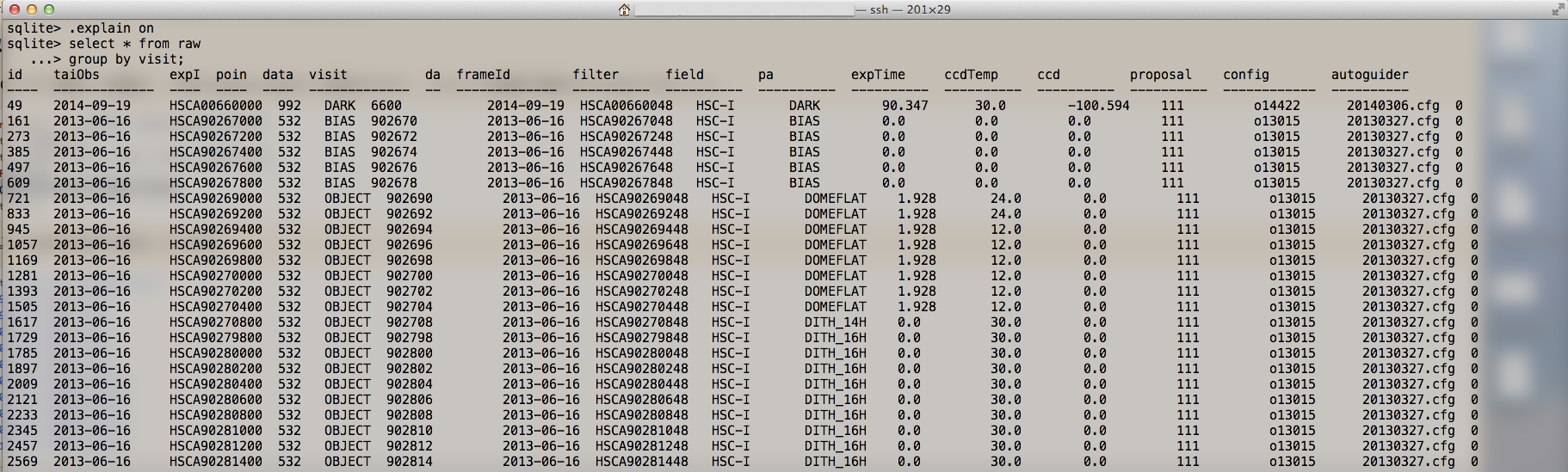
In Figure 4, you will find 112 CCDs data in all visits. Futhermore, you can get the type of raw data and their visit IDs by sqlite3 search. For example data, we have:
- DITH_16H (G-band): visit ID 903160, 903162, ... , 903188
- DITH_16H (I-band): visit ID 902798, 902800, ... , 902870
- BIAS : visit ID 902670, 902672, ... , 902678
- DARK : visit ID 006600
- FLAT (G-band) : visit ID 903036, 903038, ... , 903044
- FLAT (I-band) : visit ID 902690, 902692, ... , 902704
Warning
It is possible that HSC pipeline commands run even when data taken one shot are imperfect for all 112 CCDs. You must check if all visits have 112 CCDs.
We briefly introduce you how to retrive registry by SQL. Types of data is stored in
table. First, we check the number and types of table in a registry. For now,
there are only raw data table named raw and raw_visit. In raw table,
information of object, visit, ccd, filter etc is stored. We show all informations
as an example in Figure 5.
# a command to show an explanation of search results
sqlite> .explain on
# showing the type of table
sqlite> .table
raw raw_visit
# selecting all header information from raw table
sqlite> select * from raw
...> group by visit; # show in the order of visit ID
# search results
#
id taiObs expI poin data visit da frameId filter field pa expTime ccdTemp ccd proposal config autoguider
---- ------------- ---- ---- ---- ------------- -- ------------- ---------- ---------- ---------- ---------- ---------- ---------- ---------- ------------ ----------
49 2014-09-19 HSCA00660000 992 DARK 6600 2014-09-19 HSCA00660048 HSC-I DARK 90.347 30.0 -100.594 111 o14422 20140306.cfg 0
...
# help command to retrive a registory by sqlite3
sqlite> .help