Warp and Stack¶
Flux scale and WCS determination files are produced in mosaic.py. In the next step of stack.py, HSC pipeline warps images to the SkyMap coordinate system (warp), and combines all images together (coadd). The warp is carried out with WCS determination files produced in mosaic.py as the initial process in stack.py. As the second process, the correction of flux scale of each [visit, ccd] using flux scale files and coadd process are carried out. In order to reduce satellite tails, ghosts, and cosmic rays, outliers are clipped and median stack is adopted. Finally, catalog files are produced after sky subtraction (2048 x 2048 pixels mesh).
Since stack.py process takes a time independent of machine environment, you can perform stack.py with a batch process (see batch). Once you submit your batch job to your computer, we recommend you to work on other things.
# Warp and stack
stack.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id filter=HSC-I tract=0 --selectId visit=902798..902808:2 ccd=0..103
stack.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id filter=HSC-G tract=0 --selectId visit=903160..903168:2 ccd=0..103
# Usage:
# stack.py <a directory for data reduction> --calib=<a directory for detrend data> --rerun=<rerun name> --id filter=<filter name> tract=<tract IDs> --selectId visit=<visit IDs> ccd=<CCD IDs>
#
# Parameters:
# --id : Specify tract IDs and filter names of output data. In the example above, we set tract Id 0 and I-band filter.
# --selectId: Specify input data. In the example above, we set all [visit, ccd] IDs that we use.
Once stack.py is done, coadd images are produced as [patch].fits in $home/hsc/rerun/[rerin]/deepCoadd/[filter]/[tract] directory for hscPipe ver 3.x. You can also find warp images as warp-[filter]-[tract]-[patch]-[visit].fits in $home/hsc/rerun/[rerin]/deepCoadd/[filter]/[tract]/[patch] directory.
If you want to display all coadd images, you can use ds9 with -mosaic parameter.
# Display all coadd images with ds9
cd $home/hsc/rerun/[rerin]/deepCoadd/[filter]/[tract]
ds9 -mosaic wcs *.fits
# The ds9 calibrate a scale of coadd images in all field of views with Scale > Scope > Global bottum
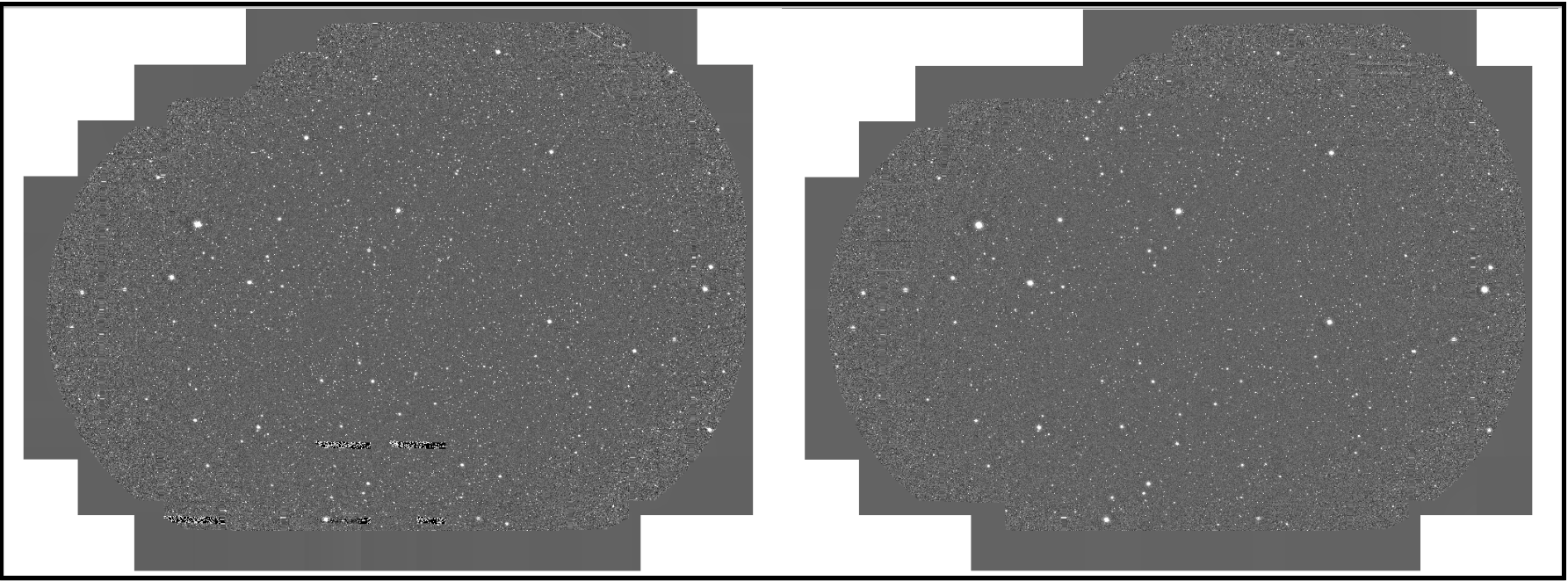
Fig 1: All coadd images of test data in I-band, and G-band, respectively. All coadd images of all data are shown in here.

Fig 2: One coadd image, a warp images of a [visit1, patch] data, and a warp images of a [visit2, patch] data.
For hscPipe ver 4.x, coadd images are created in $home/hsc/rerun/[rerun]/deepCoadd/[filter]/[tract]/[patch] as calexp-[filter]-[tract]-[patch].fits. And also, warp images, warp-[filter]-[tract]-[patch]-[visit].fits are made in the same folder.
Catalog files of detected objects are also produced during the process of stack.py. For hscPipe ver 3.x, they are found in $home/hsc/rerun/[rerin]/deepCoadd-results/[filter]/[tract]/[patch] as
- An objects catalog detected in coadd image: src-[filter]-[tract]-[patch].fits
- An objects list detected in coadd image: det-[filter]-[tract]-[patch].fits
- An objects match list between coadd image and astrometry catalog: srcMatch-[filter]-[tract]-[patch].fits
- An extension catalog file of srcMatch-[filter]-[tract]-[patch].fits: srcMatchFull-[filter]-[tract]-[patch].fits
If you want to use catalog files for your science, you can use src-[filter]-[tract]-[patch].fits. Columns in catalog files are shown in Contents of src-[filter]-[tract]-[patch].fits generated after stack.py.
For hscPipe ver 4.x, only a list of detected objects, det-[filter]-[tract]-[patch].fits is made in $home/hsc/rerun/[rerun]/deepCoadd-results/[filter]/[tract]/[patch].
You can also find sky-subtraction pattern images in the same directory as
- bkgd-[filter]-[tract]-[patch].fits
This is same in both hscPipe ver 3.x and 4.x.
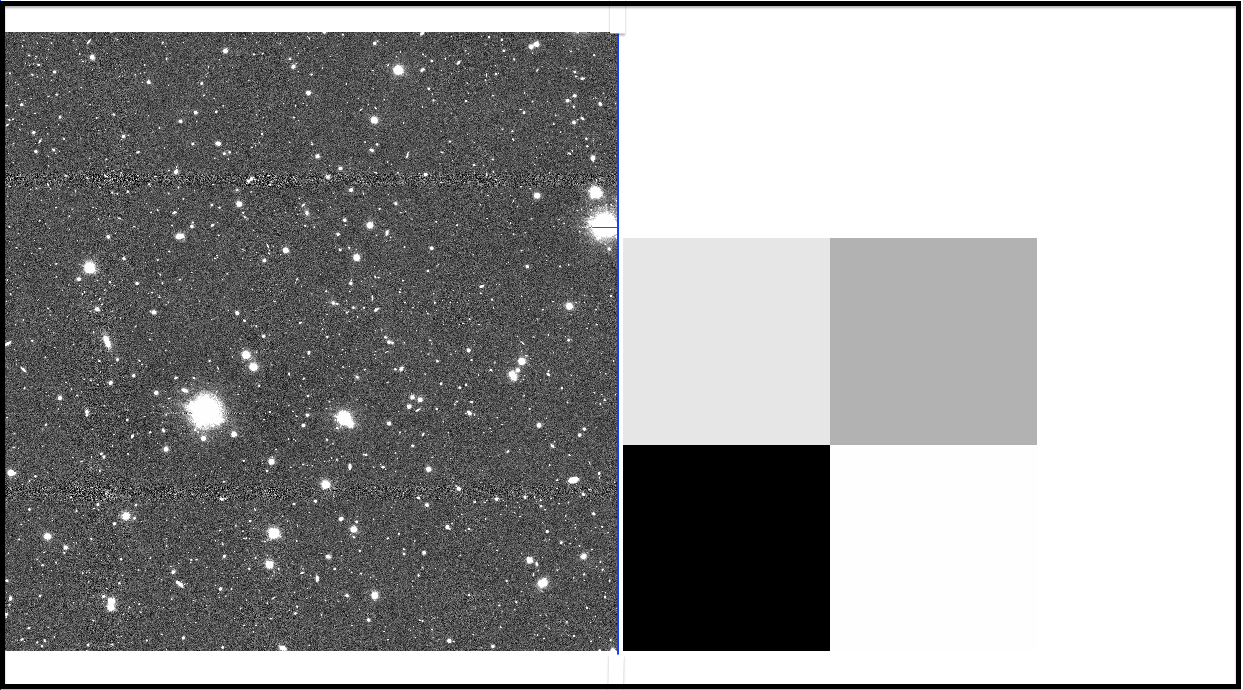
Fig 3: A coadd image of one patch and its sky-subtraction pattern image. They are arranged from left to right.
Warning
If you execute stack.py again, the data is not overwritten. So please move or delete your old data first.
The following process is only applied to hscPipe ver 3.x because the measured catalogs are created after multiband analysis in hscPipe ver 4.x. You can check the contents of catalog files by the same way of ” how to confirm a catalog file of reduceFrames.py”. It is caution that since the coordinate in centroid_sdss is referred to a pixel position in a tract, you need to correct a pixel position of [patch].fits in the tract.
# Open [patch].fits with ds9
im = afwImage.ImageF("~/hsc/rerun/dith_16h_test/deepCoadd/HSC-I/0/5,5.fits")
# Examine a position of [patch].fits in a tract
im.getXY0()
# Output on your terminal
# Point2I(19900, 19900)
# Pull up information in src-[filter]-[tract]-[patch].fits using pyfits
cat1 = pyfits.open("~/hsc/rerun/dith_16h_test/deepCoadd-results/HSC-I/0/5,5/src-HSC-I-0-5,5.fits")
xy = cat1[1].data["centroid_sdss"]
# Display objects in a catalog on ds9 (we correct the position of a patch in a tract)
for x, y in xy:
ds9.dot("o", x-19900, y-19900, size=30)
# We also display a match list.
# Pull up information in srcMatchFull-[filter]-[tract]-[patch].fits using pyfits
cat2 = pyfits.open("~/hsc/rerun/dith_16h_test/deepCoadd-results/HSC-I/0/5,5/srcMatchFull-HSC-I-0-5,5.fits")
ij = cat2[1].data["src_centroid_sdss"]
# Display objects in a match list catalog with a red circle
for i, j in ij:
ds9.dot("o", i-19900, j-19900, size=50, ctype="RED")
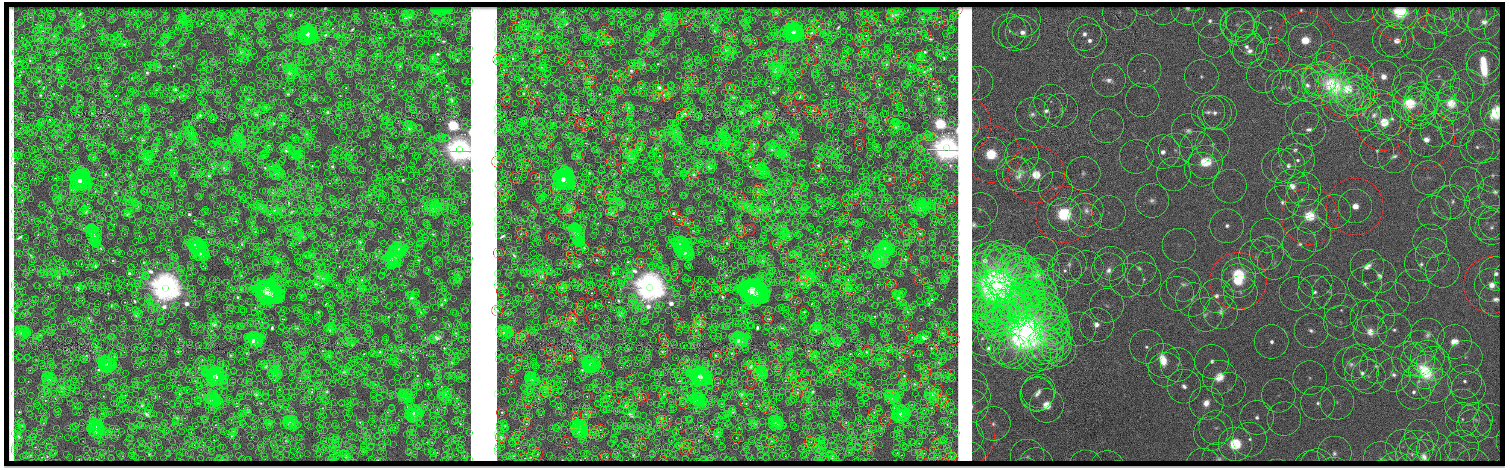
Fig 4: A coadd image with objects in src-[filter]-[tract]-[patch].fits, an image with objects in src-[filter]-[tract]-[patch].fits and srcMatchFull-[filter]-[tract]-[patch].fits, and an enlarged image are arranged from left to right.
Mask images are also included in coadd images – coadd image is [patch].fits[1], and mask image is [patch].fits[2].

