Mosaicing¶
Once you apply detrend data to each CCD, you need to define tract, and determine zero-point magnitudes and WCS.
Define tract¶
First, you need to define tract for your data with makeDiscreteSkyMap.py. In the process of makeDiscreteSkyMap.py, HSC pipeline define a square covering the entire observed sky as one tract. Since tract is determined by the entire observed sky, you must specify all data when you run makeDiscreteSkyMap.py.
Warning
If you observed multiple filters and run makeDiscreteSkyMap for each filter, the tract/patch definition in them would be slightly different. Then the multiband processing would not succeed. So please include all regions and filters in makeDiscreteSkyMap.
# Define a tract
makeDiscreteSkyMap.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id field=DITH_16H
# Usage:
# makeDiscreteSkyMap.py <a directory for data reduction> --calib=<a directory for detrend data> --rerun=<rerun name> --id field=<field name>
#
# Parameters:
# --id :Specify all objects data for the tract determination.
# In the example above, we use `field name` to specify object data.
# But you can also use `visit` or `filter`. If you want to use
# `visit` instead of `field`, you can set as follows:
# --id visit=902798..902806:2^903160..903168:2
# Note that "^" means "&", and is used when you set discrete visit number.
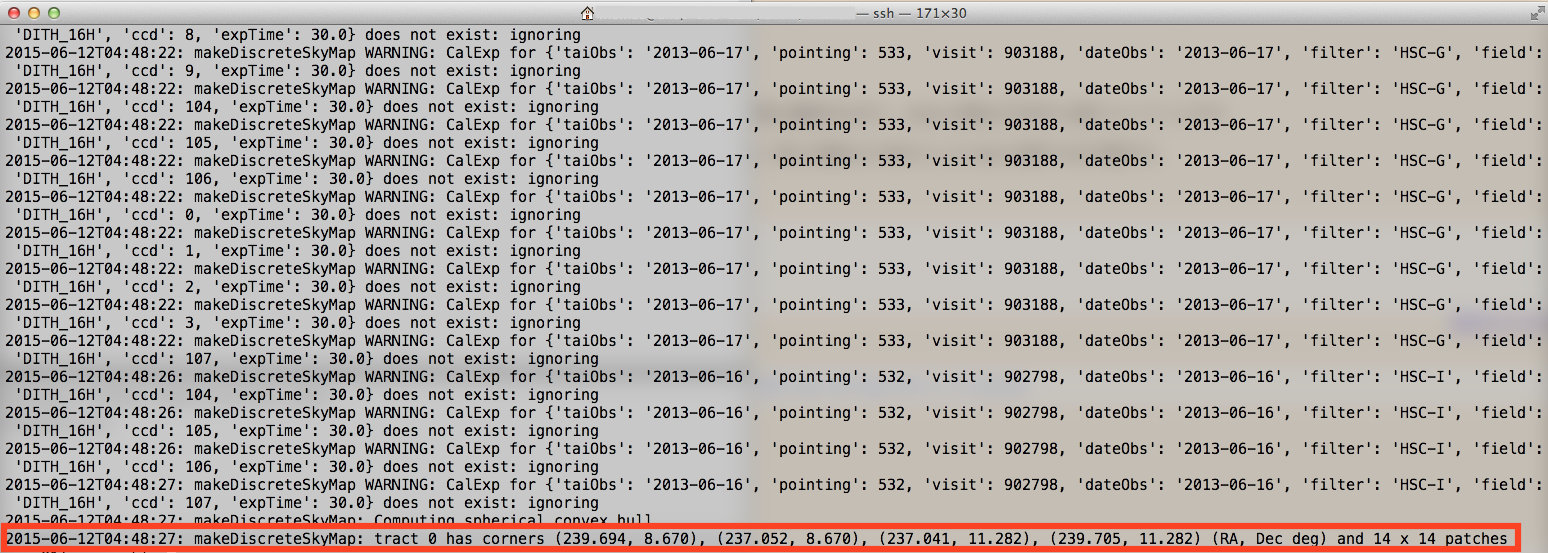
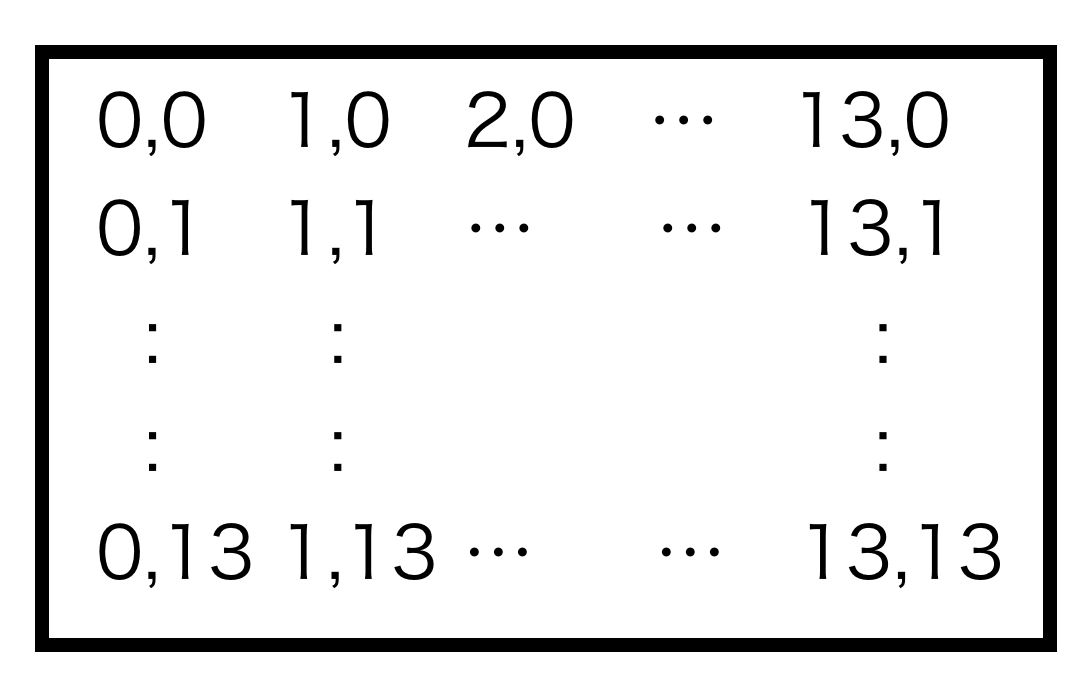
If the following message is seen on your terminal, tract setting is complete (Figure 1). In this example, tract=0 which has 14 × 14 patches is defined. The configuration of patches are presented in Figure 2.
2015-06-18T07:47:06: makeDiscreteSkyMap: Computing spherical convex hull
2015-06-18T07:47:06: makeDiscreteSkyMap: tract 0 has corners (239.635, 8.771), (236.992, 8.771), (236.981, 11.384), (239.646, 11.384) (RA, Dec deg) and 14 x 14 patches
If you need to set tract same as SSP, or tract and patch sizes manually, see Tips for tract definition.
Run mosaic.py¶
HSC pipeline command to run mosaic process is mosaic.py. mosaic.py generates a matched list of reference bright stars from each CCD in a given tract. Then it solves spatially-verying sorrection terms of relative positions and flux scales on different CCDs/visits.
# Determine zero-point magnitudes and WCS
mosaic.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id visit=902798..902808:2 ccd=0..103 tract=0
mosaic.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id visit=903160..903168:2 ccd=0..103 tract=0
# Usage:
# mosaic.py <a directory for data reduction> --calib=<a directory for detrend data> --rerun=<rerun name> --id visit=<visit ID> ccd=<CCD ID> tract=<tract ID>
#
# Parameters:
# --id :Specify all objects data for mosaic.
# In the example, we specify visit IDs of 902798, 902800, 902802, 902804, 902806, 902808,
# and CCD IDs 0 to 103 (all CCD data), and tract ID 0.
Warning
In case of narrow-band filter NB387 and NB515, please refer to here for mosaic.py execution.
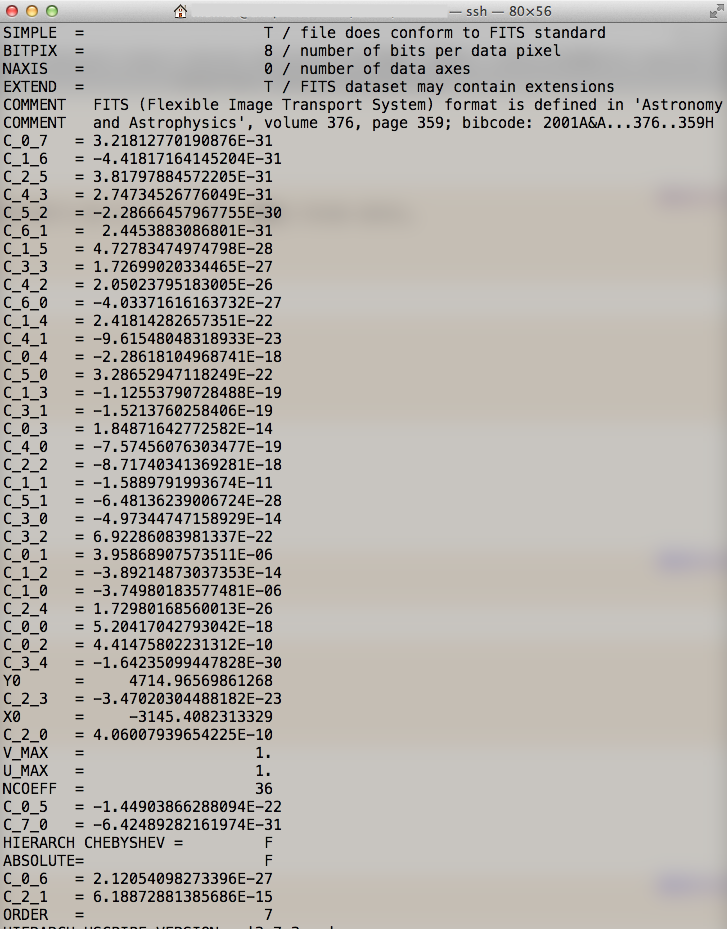
Once you complete mosaic.py, you can find fcr-[visit]-[ccd].fits and wcs-[visit]-[ccd].fits under $home/hsc/rerun/[rerun]/[pointing]/[filter]/corr/[tract] directory. These fits files are flux scale and WCS determination files, respectively. They will be applied in the process of Warp and Stack.
Scaling value of each pixel flux is evaluated with seven-order plane function and fluxmag0 in fcr-[visit]-[ccd].fits. The WCS is derived with polynomial coefficient in wcs-[visit]-[ccd].fits.
Then the results of mosaic are reflected to each CCD images and catalogs.
Note
If you analyze each CCD images and catalogs deeply, we recommend this step just after mosaic. In case that you prioritize stacked data analysis, please execute this step at the end of the procedure. Or if you do not need calibrated CCD images/catalogs, you can skip this step.
HSC pipeline command to run this process is calibrateExposure.py and calibrateCatalog.py.
# Applying the mosaic solution to the visit/CCD images
calibrateExposure.py $home/hsc --rerun=dith_16h_test --id visit=902798..902808:2 ccd=0..103 tract=0 -j 8
# Usage:
# calibrateExposure.py <a directory for data reduction> --rerun=<rerun name> --id visit=<visit ID> ccd=<CCD ID > tract=<tract> -j <number of thread>
#
# Parameters:
# --id : Specify the object data used in calibrateExposure.py で使用する天体データを指定
# -j : Specify the number of thread. In the above example -j 8 is set, then it works with 8 threads.
After calibrateExposure.py , you can find CALEXP-[visit]-[ccd].fits files in $home/hsc/rerun/[rerun]/[pointing]/[filter]/corr/0000/.
Same as above, the results of mosaic are applied to each catalogs.
# Applying the mosaic solution to the visit/CCD catalogs
calibrateCatalog.py $home/hsc --rerun=dith_16h_test --id visit=902798..902808:2 ccd=0..103 tract=0 --config doApplyCalib=False -j 8
# Parameters:
# --config doApplyCalib=False : If you do not calibrate fluxes to magnitudes, you set to False.
After completing calibrateCatalog.py, you can find CALSRC-[visit]-[ccd].fits under $home/hsc/rerun/[rerun]/[pointing]/[filter]/output/0000.
You sometimes get the error about “Not opening files” at the edge of the FOV, but you can ignore it because mosaic is not well solved there.
Evaluation results of mosaic¶
The mosaic.py can output evaluation results of mosaic process as well as flux scale and WCS determination files. Note that the command options are different between hscPipe ver 3.x and 4.x.
# Run mosaic.py with stating to output evaluation results
# hscPipe ver 3.x
mosaic.py $home/hsc --calib=$home/hsc/CALIB --rerun=dith_16h_test --id visit=902798..902808:2 ccd=0..103 tract=0 --config outputDiag=True outputDir=~/hsc/rerun/dith_16h_test/output_mosaic --clobber-config
# Usage for hscPipe ver 3.x:
# mosaic.py <a directory for data reduction> --calib=<a directory for detrend data> --rerun=<rerun name> --id visit=<visit ID> ccd=<CCD ID> tract=<tract ID> --config outputDiag=True outputDir=<a directory for evaluation results> --clobber-config
# hscPipe ver 4.x
mosaic.py $home/HSC --calib=$home/hsc/CALIB --rerun=dith_16h_test --id visit=902798..902808:2 ccd=0..103 tract=0 --diagnostics --diagDir=~/hsc/rerun/dith_16h_test/output_mosaic --clobber-config
# Usage for hscPipe ver 4.x:
# mosaic.py <a directory for data reduction> --calib=<a directory for detrend data> --rerun=<rerun name> --id visit=<visit ID> ccd=<CCD ID> tract=<tract ID> --diagnostics --diagDir=<a directory for evaluation results> --clobber-config
#
# Parameters:
# --config :Add non-default parameters.
# In this example, we set outputDiag=True outputDir=<a directory for evaluation results>,
# and output their thamnails.
# --diagnostics :Add to output evaluation results.
# --clobber-config:Verbosity for rerunning mosaic.py.
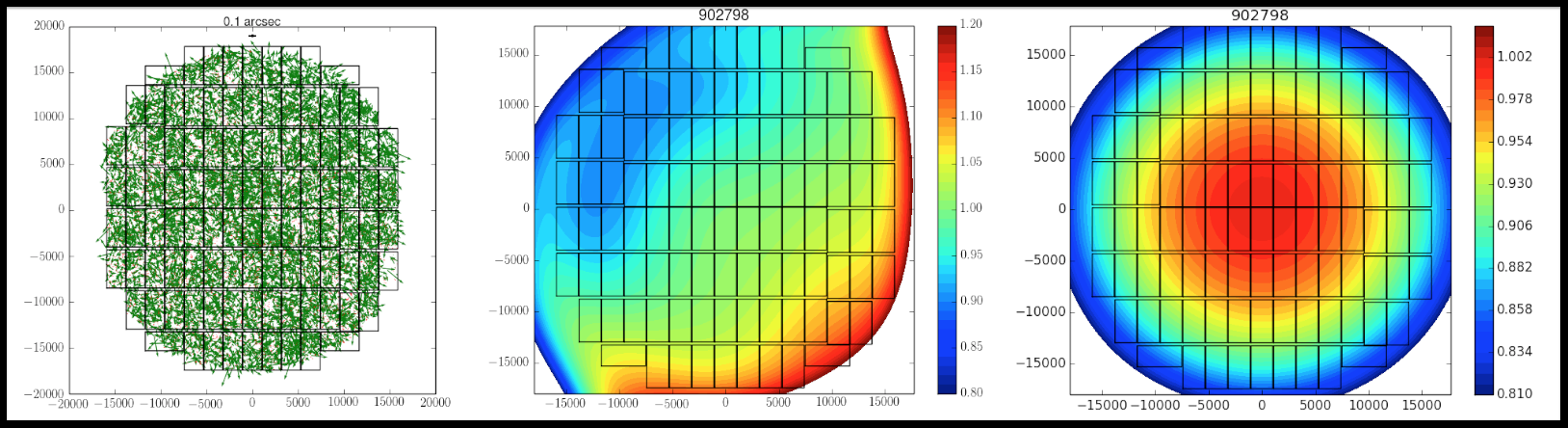
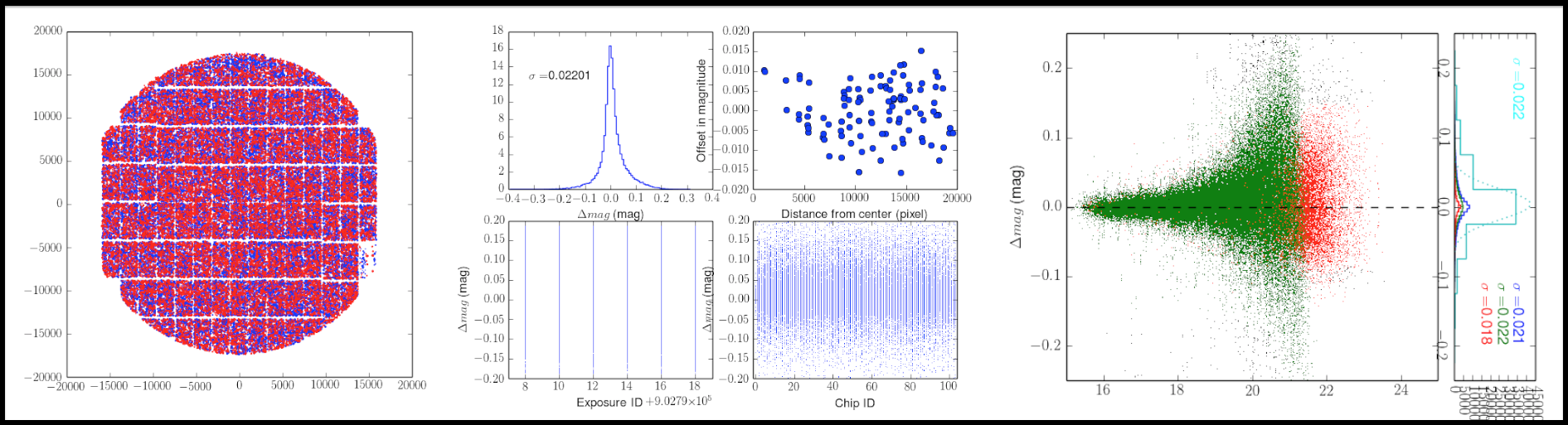
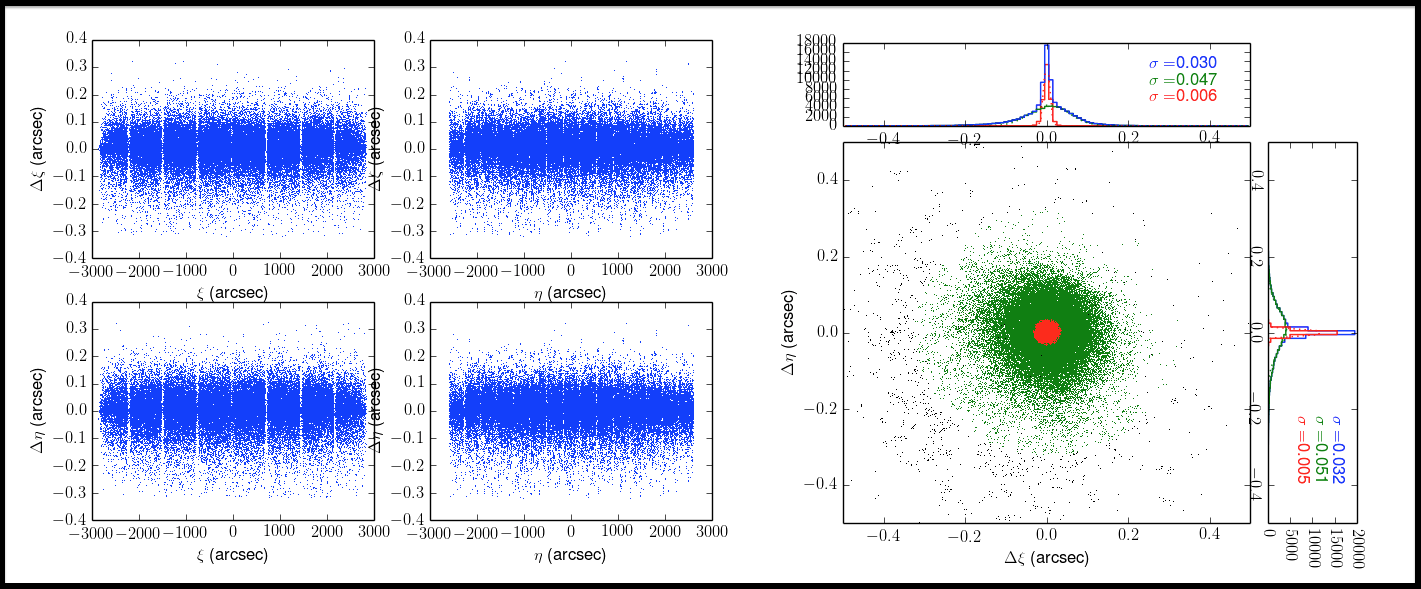
Here are evaluation results of mosaic.py.
Evaluation results from each visit
- Positional offsets: ResPosArrow2D_[visit].png
- Residuals of flux to distortion pattern: fcont_[visit].png
- Distortion pattern of reduceFlat: jcont_[visit].png (confirm that distributions is in a concentric fasion)
Evaluation results from entire visit
- Deviation from mean flux of each visit: DFlux2D.png
- Residuals of fluxes: ResFlux.png
- Deviation in magnitudes of detected sources (※): MdM.png
- Offsets of WCS: PosDPos.png
- Offsets of WCS: ResPosScatter.png
- Other outputs
- catalog.fits
- ccd.dat
- ccdScale.dat
- coeffs.dat
- dmag.dat
- dpos.dat
(※) Green and red dots denote deviation of catalogs in HSC pipeline from astrometry catalog, and deviation from the mean of all visit in sources which are not in MATCH catalog