Making data for detrending¶
For detrending and calibrating each CCD data, Bias, Dark, Flat, Fringe, and Sky data are required. The analysis is carried out in order of making Bias, Dark, Flat, Fringe, and Sky. Note that Fringe data is usually needed for the analysis in y-band, NB0816, NB0921, NB0926, IB0945, NB0973 and NB1010.
Note
lossless FITS tile compression is now the default for data products written by the butler. For details on how to configure this behavior (including disabling compression), see https://community.lsst.org/t/fits-tile-compression-support/2233. Please use the latest version of tools for fits (which also includes the DS9).
Making Bias data¶
Bias data is generated by removing an overscan from 0 set integration data (this is called as bias raw data). Each CH of one CCD has a 16-pixels’ overscan region at the left or right edge of it (refer to HSC CCDs). Pipeline evaluates the mean value along the shortest axis of the overscan region, fits spline to the longest axis, and subtract this value from bias raw data. Finally the Bias data are produced by averaging the value across all visits after removing some outliers from these data. This can be done by hscPipe command constructBias.py.
# Making Bias data
# In case of data reduction directory is ~/HSC, directory of data for detrending ~/HSC/CALIB, and rerun name calib。
# Please note that; if there is no directory named ~/HSC/CALIB, the data for detrending are created in ~/HSC/rerun/[rerun].
constructBias.py ~/HSC --calib ~/HSC/CALIB --rerun calib --id visit=18624..18632:2 --batch-type=smp --cores=4
# constructBias.py [data reduction directory] --calib [directory of data for detrending] --rerun [rerun name] --id [visit]
# Options
# --calib: Name of a directory of data for detrending. This must match the regular expression [A-Za-z0-9_+-]+.
# --rerun: Rerun name
# --id: Input data ID. Here, specify the data ID of bias raw data. "visit=18624..18632:2" means specifying visit ID from 18624 to 18632 with interval 2.
# You can also use "^" when you specify target visits like "visit=18624^18630".
# You can exclude non-scientific CCDs (DETID=>104) by setting ccd=0..103 (not necessarily needed).
# If your fits name is later than HSCA13880000.fits, exclude DETID9 by setting ccd=0..8^10..103.
# ccd: DETID9 is excluded because it stops read-out because of defects.
# If you do not exclude DETID9, the procedure ConstructFlat.py or later may not be completed.
# Or, you can use the column names in the table of registry.sqlite3 (e.g., field=BIAS).
# --batch-type [slurm, pbs, smp]: Specify the batch system you use.
# --cores: Number of cores (Slurm/SMP only).
You can use batch system (on which the jobs are submitted to job control system) for all commands of making data for detrending in hscPipe. It provides batch process options depending on your machine environments. Detailed information could be checked with –help option.
# Running processes in local machine (using 4 cores)
--batch-type=smp --cores=4
# Submitting a batch script to another machine(using 2 nodes, 2 processes, total 4 threads)
--nodes=2 --cores=2
Note
If you want to save standard and standard error output as one file, add “2>&1 | tee logfile” to the end of each command. This log file is sometimes needed for investigation by Helpdesk.
Warning
HSC data analysis machine for open use(hanaco)does not have job control system. So please add the option in case of “Running processes in local machine”.
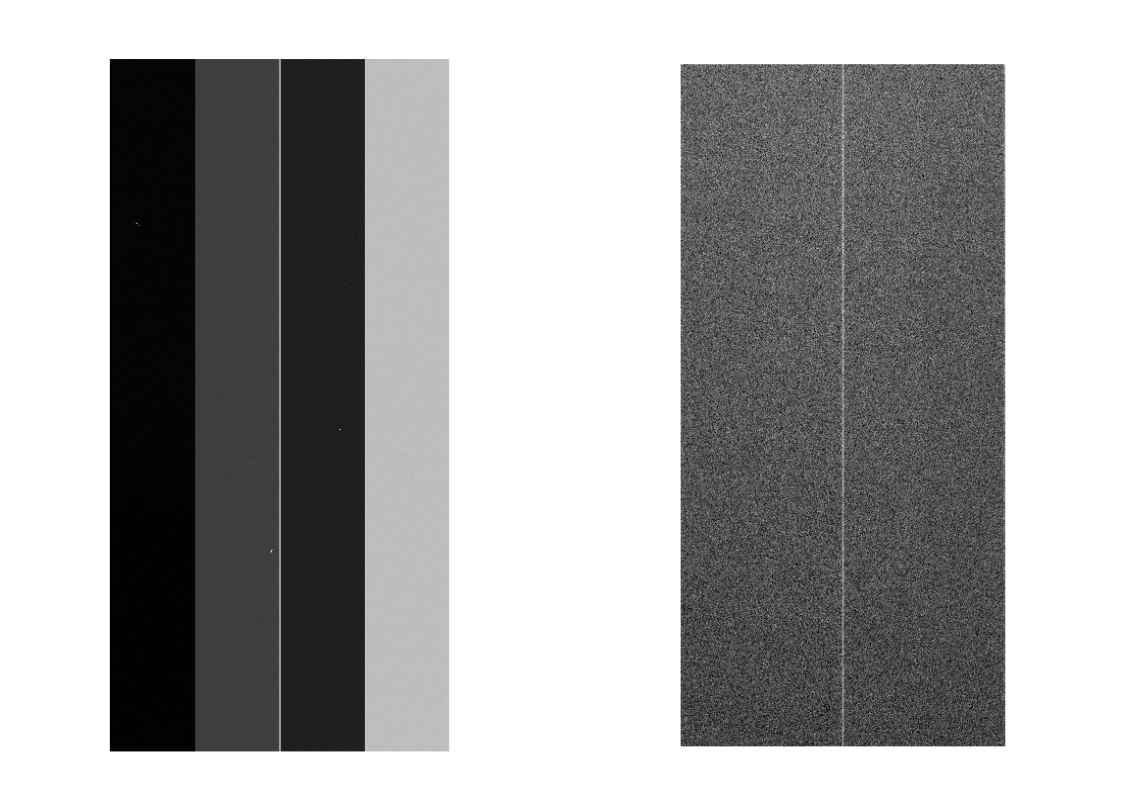
The Bias data are created as ~/HSC/rerun/calib/BIAS/[dateObs]/NONE/BIAS-[dateObs]-[ccd].fits, and other intermediate ones are in ~/HSC/rerun/[rerun]. Figure 1 shows an example of bias raw data and Bias data. The later is smaller than former because of cutting overscan and blanc regions in a CCD.
Then make a registry. From hscPipe5, the processing progresses without registering data for detrending. However, the data loading becomes faster with registry when you use a number of cores. We recommend that you register the data for detrending.
# Register the data for detrending
ingestCalibs.py ~/HSC --calib ~/HSC/CALIB/ '~/HSC/rerun/calib/BIAS/*/*/BIAS-*.fits' --validity 30
# ingestCalibs.py [data reduction directory] --calib [directory of data for detrending] '[fits file name]' --validity [days] --mode=link
# Option
# --validity: Calibration validity period (days)
# --mode: Mode of delivering the files to their destination.
# If you choose "link", the link to raw data is created.
There is a registry “calibRegistry.sqlite3” in ~/HSC/CALIB. This file can also be checked by SQLite.
# Load registry by SQLite
sqlite3 calibRegistry.sqlite3
# Read header information
sqlite> .header on
# Show tables
sqlite> .tables
bias dark flat fringe sky
bias_visit dark_visit flat_visit fringe_visit sky_visit
# Registered Bias data are in the bias table.
sqlite> select * from bias;
id|filter|ccd|validStart|calibDate|validEnd
1|NONE|60|2014-12-19|2015-01-18|2015-02-17
2|NONE|99|2014-12-19|2015-01-18|2015-02-17
3|NONE|85|2014-12-19|2015-01-18|2015-02-17
4|NONE|23|2014-12-19|2015-01-18|2015-02-17
5|NONE|35|2014-12-19|2015-01-18|2015-02-17
6|NONE|32|2014-12-19|2015-01-18|2015-02-17
7|NONE|102|2014-12-19|2015-01-18|2015-02-17
8|NONE|74|2014-12-19|2015-01-18|2015-02-17
...
# Check the number of CCDs and compare validStart/validEnd with the period specified at --validity.
# The other tables are empty.
Making Dark data¶
Dark data is used to correct dark current in a CCD stored within any integration time. It is generated from dark raw data by subtracting an overscan and Bias data. In the hscPipe processing, the intermediate data which subtracted average counts of overscan pixels from each dark raw data is created. Then the final Dark data is generated from the data which subtracted Bias from the average of the intermediate data. The hscPipe command constructDark.py is used for this step. You can use batch system when you execute this command.
# Making Dark data
constructDark.py ~/HSC --calib ~/HSC/CALIB --rerun calib --id visit=18336 --batch-type=smp --cores=4
# constructDark.py [data reduction directory] --calib [directory of data for detrending] --rerun [rerun name] --id [visit, field, ...]
# The options are the same with constructBias.py
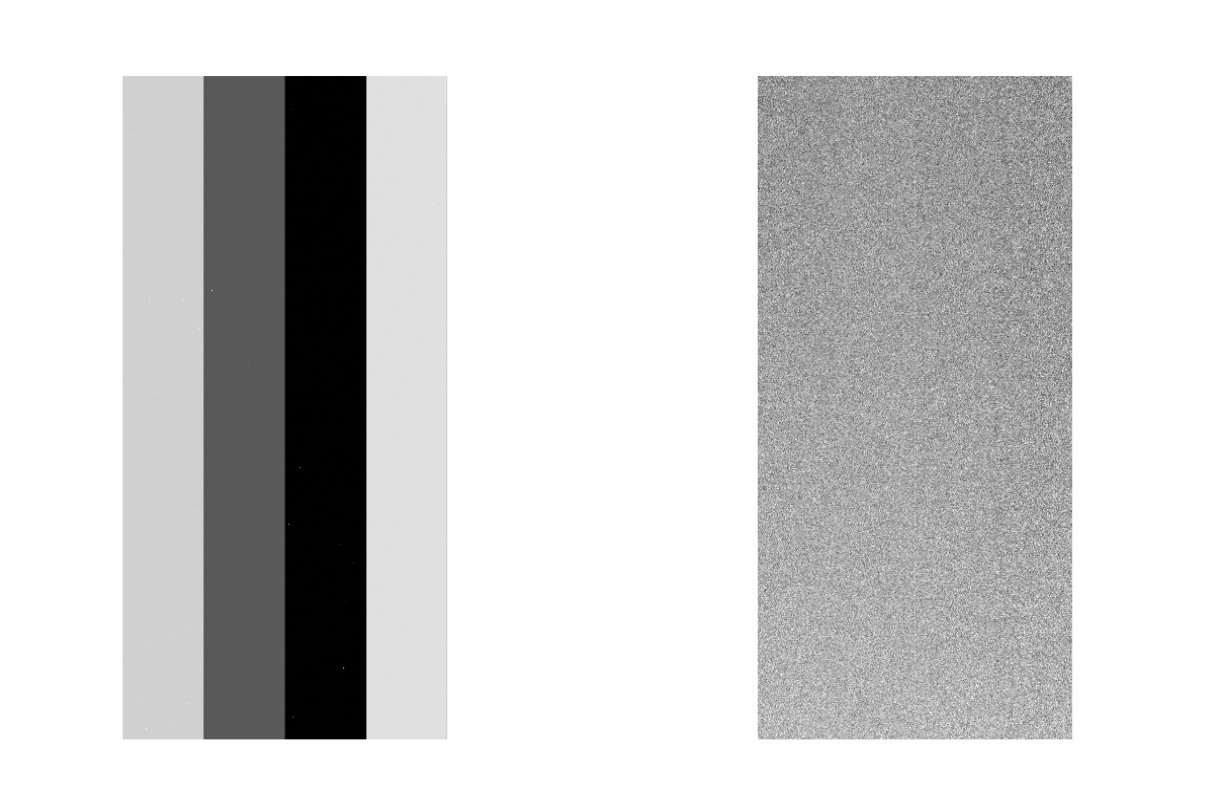
The Dark data is output as ~/HSC/rerun/calib/DARK/[dateObs]/NONE/DARK-[dateObs]-[ccd].fits. The intermediate files are in ~/HSC/rerun/[rerun]. Figure 2 shows dark raw and generated Dark data. There is no overscan and blank regions, so the Dark data is smaller than original raw data.
Then register the Dark data.
# Register the Dark data
# Options are the same as Bias data.
ingestCalibs.py ~/HSC --calib ~/HSC/CALIB/ '~/HSC/rerun/calib/DARK/*/*/DARK-*.fits' --validity 30 --mode=link
Making Flat data¶
There are two types of flatfielding; dome flat which uses the observed data of an illuminated screen or wall, and sky flat which made from on-source sky data. Basically the dome flat is used for HSC observation, so we describe how to create dome flat data in this section. The sky flat data can be made from the same command.
To begin with, the mean value of the overscan is subtracted from flat raw data of each [visit, CCD]. Then the final Flat data is created by subtracting Bias and Dark data from the data made in the previous step. If you do not execute Bias and Dark subtraction, this process is skipped. It can be done by constructFlat.py. Batch system is also available for the command.
Note
Creating Bias and Dark data can be skipped if you want. However Flat data is neseccary.
# Making Flat data
constructFlat.py ~/HSC --calib ~/HSC/CALIB/ --rerun calib --id visit=17636..17646:2 --batch-type=smp --cores=4
# constructFlat.py [data reduction directory] --calib [directory of data for detrending] --rerun [rerun name] --id [visit, field, filter...]
# Note that Flat data should be created for each filter.
# In case of sky flat, you have to specify sky flat raw data (about 30 shots) at --id.
# Option
# --config isr.doBias=False isr.doDark=False: If you skip creating Bias and Dark data, add this option.
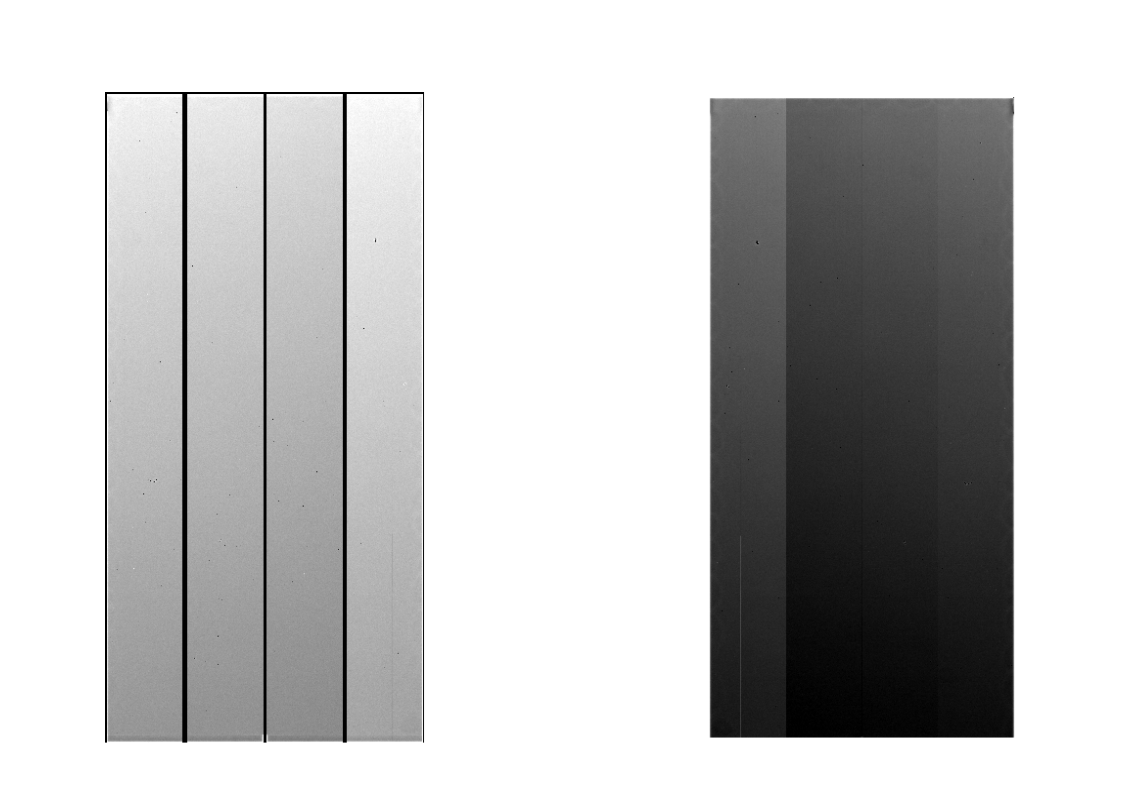
Generated Flat data files are ~/HSC/rerun/calib/FLAT/[dateObs]/[filter]/FLAT-[dateObs]-[filter]-[ccd].fits, and the other intermediate ones are in ~/HSC/rerun/[rerun]. The Flat and flat raw images are shown in figure 3.
Then add the Flat data to calibRegistry.sqlite3 same as Bias and Dark.
# Register Flat data
# Options are the same as Bias data.
ingestCalibs.py ~/HSC --calib ~/HSC/CALIB/ '~/HSC/rerun/calib/FLAT/*/*/FLAT-*.fits' --validity 30 --mode=link
Making Fringe data¶
Fringe is the pattern caused by interference between surface of CCD and it becomes obviously in longer wavelength. The pipeline supports y-band, NB0816, NB0921, NB0926, IB0945, NB0973 and NB1010 by default. The Fringe data is made from the object images when you execute constructFringe.py command in hscPipe.
# Making Fringe data
constructFringe.py ~/HSC --calib ~/HSC/CALIB --rerun calib --id visit=18480..18490:2 --batch-type=smp --cores=4
# constructFringe.py [data reduction directory] --calib [directory of data for detrending] --rerun [rerun name] --id [visit, field, filter..]
# Options are the same as Bias.
# Specify y-band, NB0921, NB0926, or NB0973 object data as the input.
# Option
# --config isr.doBias=False isr.doDark=False: If you skip creating Bias and Dark data, add this option.
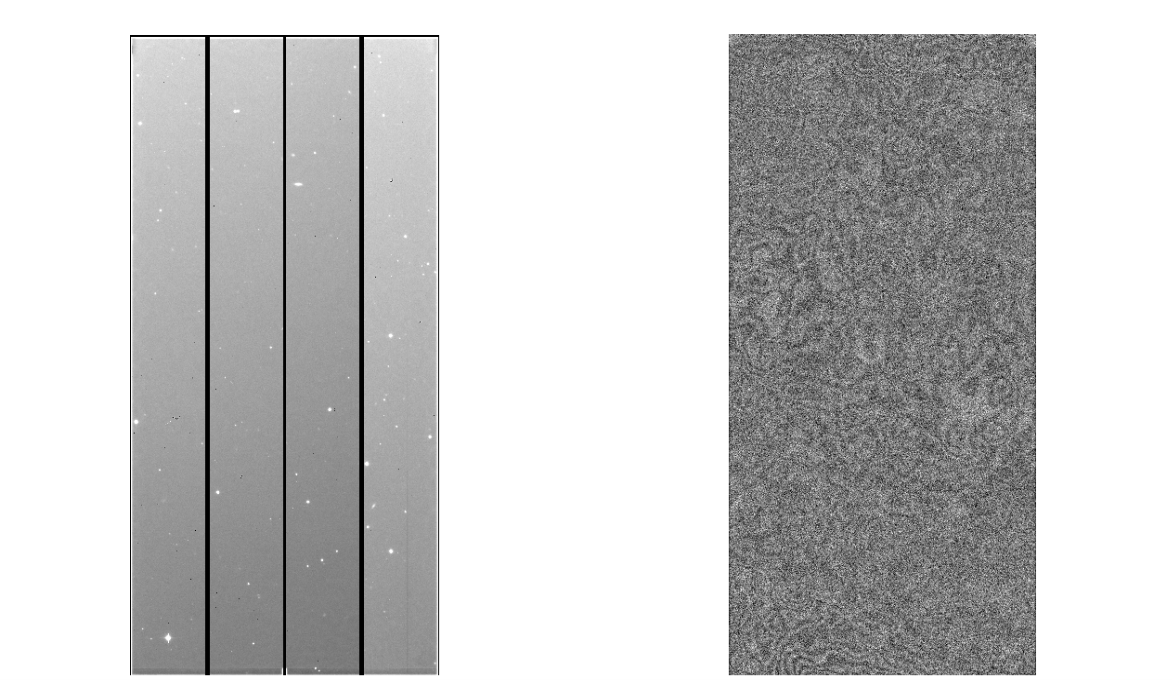
The created Fringe data are ~/HSC/rerun/calib/FRINGE/[dataObs]/[filter]/FRINGE-[dateObs]-[filter]-[ccd].fits, the intermediate files are output in ~/HSC/rerun/[rerun]. The object raw and Fringe image are shown in figure 4.
Fringe data have to be also registered.
# Register Fringe data
ingestCalibs.py ~/HSC --calib ~/HSC/CALIB/ '~/HSC/rerun/calib/FRINGE/*/*/FRINGE-*.fits' --validity 30 --mode=link
Making Sky data¶
This is an implementation of the sky subtraction algorithm advocated by the Hyper Suprime-Cam team at NAOJ. There was the over sky subtraction problem around bright objects in hscpipe 5 or earlier. From hscPipe 6, hscpipeline generates SKY images to correct the over sky subtarction around bright objects (Description of global sky subtraction). The Sky data is made from the object images when you execute constructSky.py command in hscPipe.
# Making Sky data
constructSky.py ~/HSC --calib ~/HSC/CALIB --rerun calib --id visit=18480..18490:2 --batch-type=smp --cores=4
# constructSky.py [data reduction directory] --calib [directory of data for detrending] --rerun [rerun name] --id [visit, field, filter..]
# Options are the same as Bias.
# Option
# --config isr.doBias=False isr.doDark=False: If you skip creating Bias and Dark data, add this option.
The created Sky data are ~/HSC/rerun/calib/SKY/[dataObs]/[filter]/SKY-[dateObs]-[filter]-[ccd].fits, the intermediate files are output in ~/HSC/rerun/[rerun].
Sky data have to be also registered.
# Register Sky data
ingestCalibs.py ~/HSC --calib ~/HSC/CALIB '~/HSC/rerun/calib/SKY/*/*/SKY-*.fits' --validity 30 --mode=link